Heterotopic ossification (HO) is a standard illness occurred in smooth tissues after harm. The current examine utilized the bioinformatics methodology to investigate the HO samples in a mouse burn/tenotomy-induced HO mannequin to establish the doable key factors and therapy targets.
MethodsThe transcriptome profiles of GSE126118 had been obtained from the Gene Expression Omnibus (GEO) database.
The examine was based mostly on a mouse burn/tenotomy-induced HO mannequin, and a couple of tenotomy samples and three unhurt contralateral hindlimb tendon samples had been collected at three weeks after harm for additional evaluation.
The transcripts per million method was carried out for background correction and normalization; then, the differentially expressed genes (DEGs) had been detected utilizing the limma R package deal with the settings p < 0.01 and ∣log2FC∣ > 2.0.
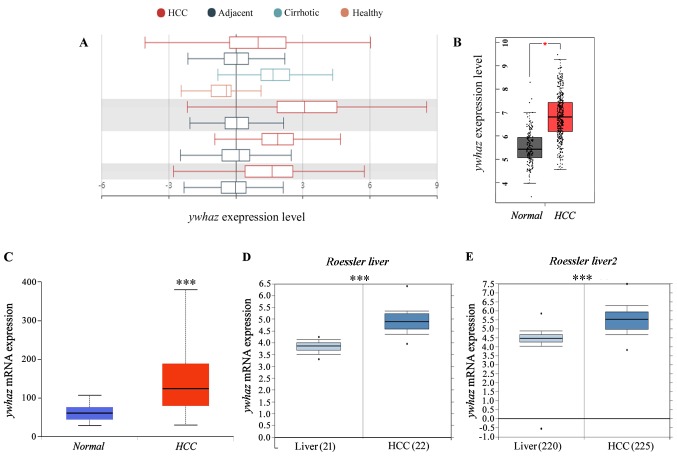
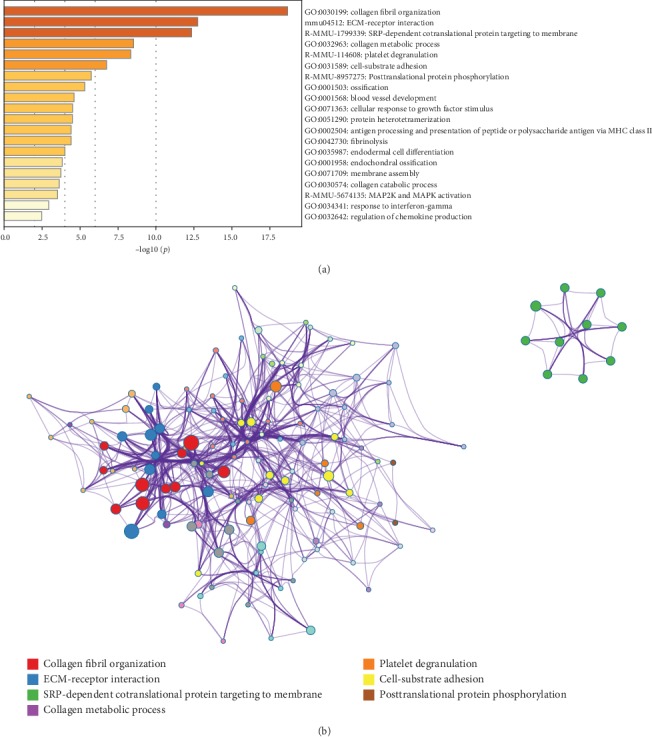
The Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment, and the protein–protein interplay (PPI) community evaluation had been carried out with the detected DEGs.A complete of 74 DEGs had been upregulated, and 159 DEGs had been downregulated between the tenotomy and unhurt tendon group.
Pathway and course of enrichment analyses demonstrated that the upregulated DEGs had been primarily related to phrases associated to ECM transforming, ossification, angiogenesis, irritation, and many others., and the downregulated DEGs had been primarily related to oxidative phosphorylation, metabolic course of, and many others.
The outcomes of GO, KEGG, and PPI community analyses advised that the ECM transforming, ossification, angiogenesis, and irritation processes had been markedly upregulated in the tenotomy website. And the oxidative phosphorylation and metabolic processes had been markedly downregulated. These findings present helpful clues for highlighting the traits of late-stage HO and investigating doable remedies.
Functional Analysis of Differentially Expressed Acetylated Spermatozoal Proteins in Infertile Men with Unilateral and Bilateral Varicocele.
Sperm proteins bear post-translational modifications, similar to phosphorylation, acetylation, and ubiquitination, which in flip play a key function in figuring out their fertilizing capacity. In the present examine, we examined the sperm proteome of males with unilateral and bilateral varicocele to establish the key proteins affected by acetylation to realize an perception into the distinction in the severity of affected sperm operate in the latter.
An LTQ-Orbitrap Elite hybrid mass spectrometer system was used to profile the sperm proteome in pooled unilateral and bilateral varicocele sufferers.
Bioinformatics database and instruments, similar to UniProtKB, Ingenuity Pathway Analysis Software (IPA) and Metacore, had been used to establish the differentially expressed proteins (DEPs) concerned in the acetylation course of. A complete of 135 DEPs in the spermatozoa of unilateral and bilateral varicocele sufferers had been discovered to be affected by acetylation. The majority of these DEPs discovered had been regulated by key transcription elements similar to androgen receptor, p53, and NRF2.
Furthermore, the DEPs predicted to be affected by the acetylation course of had been related to fertilization, acrosome response, mitochondrial dysfunction and oxidative stress.
Aberrant expression of proteins and their differential acetylation course of might have an effect on the regular physiological features of spermatozoa. Protein–protein interactions recognized dysregulation of the proteasome complicated in the bilateral varicocele group.
Damage to the proteasome complicated might end in aggregation of the misfolded proteins, which in flip improve sperm DNA injury and apoptosis in sufferers with bilateral varicocele.